
——Guidelines for the establishment of the China Academy of Advanced Science and Technology

——Guidelines for the establishment of the China Academy of Advanced Science and Technology



DeepETPicker only requires a small amount of manual annotation of particles for training, and can achieve fast and accurate 3D particle automatic selection. In order to reduce the need for manual annotation, DeepETPicker prefers simplified labels to replace real labels, and adopts a more efficient model architecture, richer data augmentation techniques, and overlapping partitioning strategies to improve the performance of the model when the training set is small; In order to improve the speed of particle localization, DeepETPicker adopts GPU accelerated average pooling non maximum suppression post-processing operation, which improves the selection speed dozens of times compared to existing clustering post-processing methods. For the convenience of users, the team has launched open-source software with simple operation and user-friendly interface to assist users in image preprocessing, particle annotation, model training and inference, and other operations.
The overall workflow of using DeepETPicker to select particles in frozen electron tomography images by researchers includes training and inference stages. In the preparation stage of training data, weak label TBall-M was selected to replace the real mask to reduce the burden of manual annotation. In terms of model architecture design, research introduces coordinate convolution and image pyramid to 3D ResUNet segmentation architecture to improve localization accuracy. In the model inference stage, DeepETPicker adopts an overlapping fault map partitioning strategy to avoid the negative impact caused by poor edge voxel segmentation accuracy, and then combines MP-NPMS operation to accelerate the particle center localization process.
This study compared the performance of DeepETPicker with the currently best particle selection method on multiple frozen electron tomography datasets, and comprehensively evaluated the quality of particle selection using six quantitative indicators. The results show that DeepETPicker can achieve fast and accurate particle selection on both simulated and real datasets, and its overall performance is superior to other existing methods; The resolution obtained from the reconstruction of biological macromolecular structures reaches the same level as using manually selected particles by experts for structural reconstruction. This reflects the practical value of DeepETPicker in in-situ high-resolution structural analysis. DeepETPicker is expected to provide support for in situ structural biology research using in situ cryoelectron microscopy technology.
The research work was supported by the Chinese Academy of Sciences' strategic leading science and technology project (Class B), the National Natural Science Foundation of China, and the national key research and development plan. The relevant technology has been granted Chinese invention patent authorization.
Paper link:论文链接
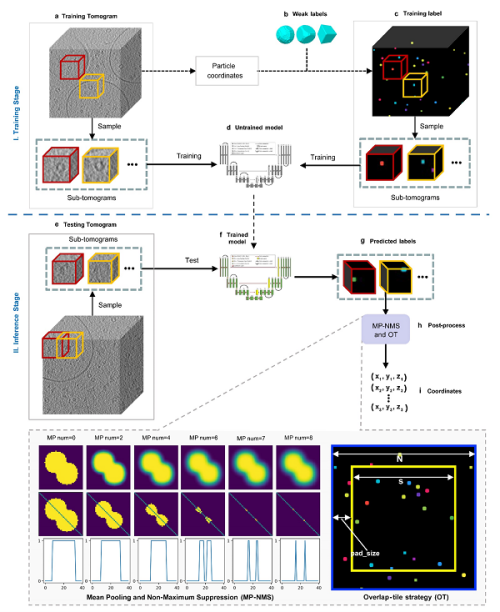
The overall workflow of using DeepETPicker to select particles in frozen electron tomography images
Zhongke Frontier(Xiamen)Science and Technology Research Institute©All rights reserved
Service Customer Service:4006 285 158 Postal Code:361006
Address:Science City Zhongke Building,Huangpu District,Guangzhou City
396 Jiahe Road,Huli District,Xiamen City
Website:http://www.zk-yjy.com