Avocado is an evergreen tree of the Lauraceae family, native to Central America. In order to interpret the genetic information of avocado, the Plant Genome Evolution and Secondary Metabolism Research Group of Xishuangbanna Tropical Botanical Garden, Chinese Academy of Sciences used NGS, PacBio HiFi, ONT, Pore-C and other sequencing technologies to sequence the whole genome of avocado of West Indian varieties, and assembled 841.6Mb gapless genome sequence of avocado.
Research has shown that avocados from West Indian varieties contain 40629 protein coding genes, and 12 chromosome specific centromere sequences (CSCRs) were predicted using iterative recognition and clustering methods. These CSCR sequences have complex structures and contain multiple types of transposons. Researchers further validated the accuracy of all predicted centromere, telomere, and nucleolar tissue regions in the avocado genome sequence using fluorescence in situ hybridization technology. In addition, the study identified 376 NLR genes related to disease resistance and 128 genes related to fatty acid biosynthesis. Among them, during the formation stage of triacylglycerol, the FAD2 gene is specifically expressed in fruits, providing new insights into the biosynthesis of fatty acids in avocado.
The relevant research results, titled A teleme to teleme gap free reference gene assembly of horticulture provides useful resources for identifying genes related to fatty acid biosynthesis and disease resistance, were published in Horticultural Research. The research work has received support from the National Natural Science Foundation of China, Yunnan Postdoctoral Science Foundation, and Yunnan Yunling Scholar Training Project.
Paper link
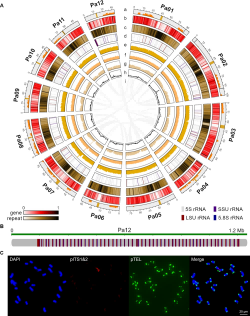
Panoramic view of genome assembly from telomere to telomere without gap in avocado




