On May 21, Guo De'an Research Group of Chinese Academy of Sciences Shanghai Institute of Materia Medica, Ma Xiaochi and Wang Chao Research Group of Dalian Medical University, together with Francis Martin, professor of French Academy of Agricultural Sciences, published a research paper entitled A genetic compatibility of cultivated human gut fungi characteristics the gut mycobiome and its relevance to common diseases online on Cell.
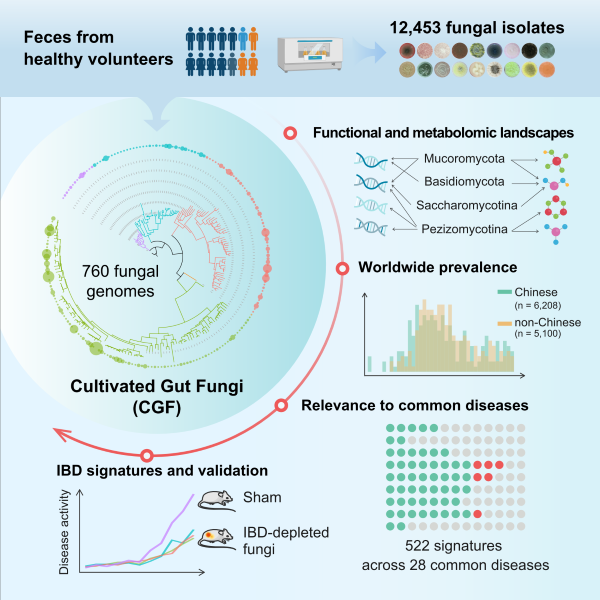
This study established the largest reference genome catalog of human cultured gut fungi through large-scale sequencing of human gut fungal cultures, completed metabolic function analysis, revealed the diversity of human gut fungi and their potential roles in common diseases, expanded existing gut fungal genome resources, and provided important reference data for the study of gut fungal community structure, biological function, and other aspects.
The human digestive system is composed of various microorganisms, including bacteria, archaea, fungi, and viruses. At present, there have been many studies on the community structure and biological functions of human intestinal bacteria. As an important component of the gut microbiota, the gut fungal community can have an impact on various physiological and pathological processes of the host. Although researchers are able to study the gut fungal community through the DNA meta barcodes of fungal 18S rRNA genes or ribosomal DNA internal transcriptional spacer regions, which enriches our understanding of the human gut fungal community, there are still limitations to systematic research on the genetics and function of human gut fungi. The reason for the above phenomenon is due to the limited reference genome.
This study used culture group technology to cultivate 12453 fungal strains from 135 healthy Chinese fecal samples and performed whole genome sequencing, obtaining 760 gut fungal genomes (covering 48 families and 206 species), of which 69 were identified for the first time. The study conducted functional gene analysis and targeted metabolomics measurements on 206 fungi, elucidating the characteristics of gut fungi in terms of gene function and metabolites. Furthermore, the study analyzed 11000 human fecal metagenomic data, revealing the structural characteristics of gut fungal communities in both Chinese and non Chinese populations. The study analyzed the gut fungal characteristics of 28 diseases, identified disease-related gut fungal characteristic signals, and focused on the gut fungal characteristics of patients with inflammatory bowel disease. The biological functions, some active substances, and molecular mechanisms of the target strain were validated in various mouse colitis models.
The fungal genome directory constructed in this study and the functional analysis work carried out have expanded scientists' understanding of the biodiversity and disease relevance of human gut microbiota, providing new ideas for the diagnosis and treatment of gut microbiota related diseases.
Paper link
Scientists analyze the human gut fungal community and its biological functions




